Shedding new light on abnormal protein synthesis in neurodegenerative disorders
Researchers identify molecular factors that regulate aberrant protein translation in inherited amyotrophic lateral sclerosis and frontotemporal dementia
Translation factors eIF1A and eIF5B are key repressors of an abnormal protein translation process linked to neurodegenerative disorders, as reported by researchers from Science Tokyo. Using a human cell-free translation system, they reconstructed the aberrant translation of a mutated C9orf72 gene. This translation process revealed that the initiation factors (eIF1A and eIF1B) act at distinct checkpoints to suppress toxic protein synthesis implicated in frontotemporal dementia and amyotrophic lateral sclerosis.
A Deep Look into RAN Translation Regulation in Neurodegenerative Diseases
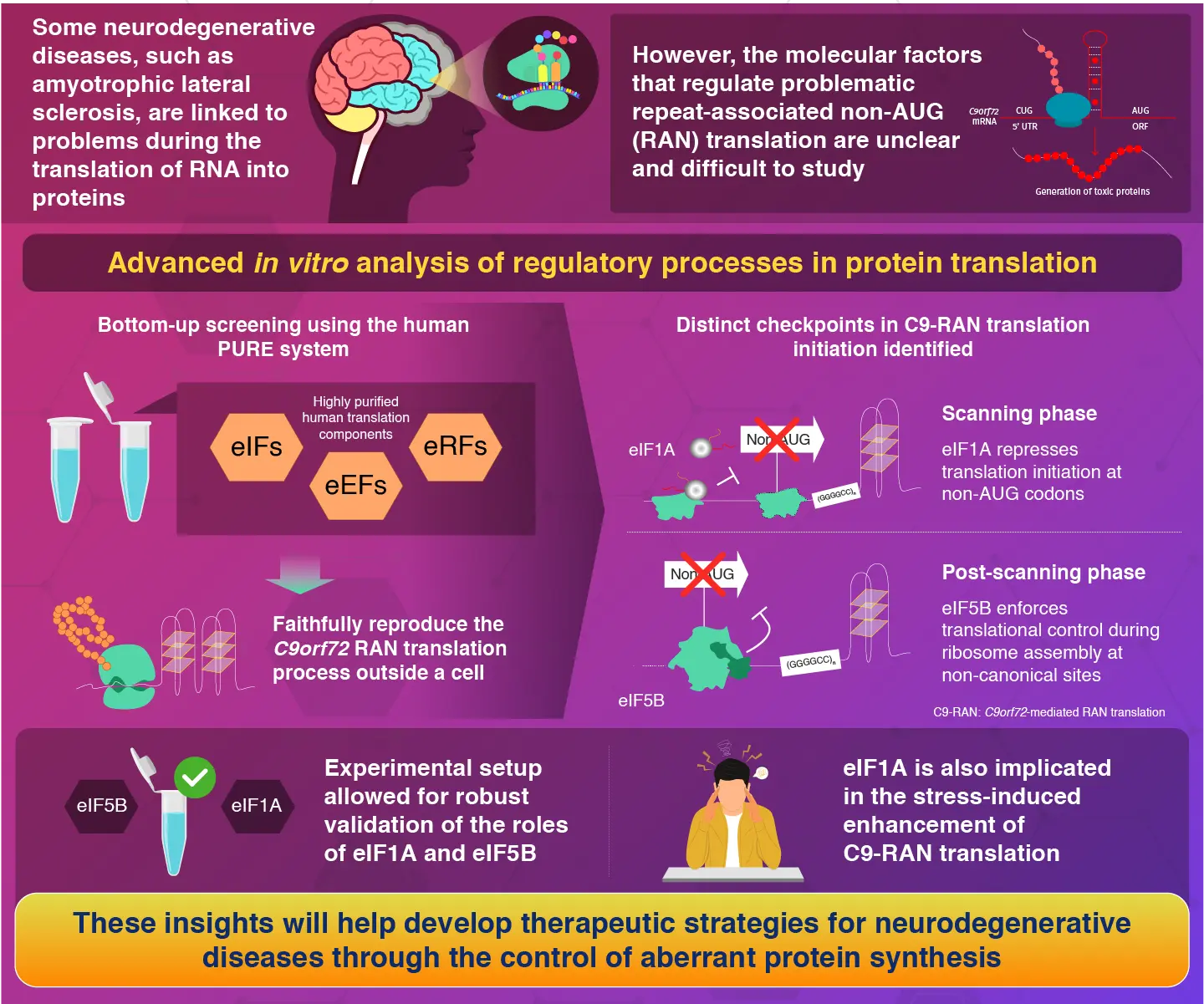
Neurodegenerative disorders, such as amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), remain a major unsolved challenge in modern medicine. While their causes can vary and are far from being fully understood, the abnormal repetition of DNA sequences is a major culprit in many inherited cases. Such genetic defects, seen in genes like C9orf72, can trigger an aberrant cellular process called repeat-associated non-AUG (RAN) translation, which defies the standard rules of protein synthesis. Instead of starting at an AUG codon—the conventional starting point—this process initiates from non-AUG codons, leading to the production of toxic proteins that accumulate and directly contribute to neuronal damage.
Despite our knowledge of why these toxic proteins exist, the specific regulators of RAN translation remain elusive. Standard research methods, which rely on whole living cells, are usually affected by indirect cellular responses that obscure the results and the effects of translation factors. This limitation has kept scientists away from a more comprehensive understanding of how the cell’s protein-making machinery is corrupted by RAN translation.
Against this backdrop, a research team led by Professor Hideki Taguchi of the Cell Biology Center, Institute of Science Tokyo (Science Tokyo), Japan, employed a bottom-up approach to identify these key regulators. Their paper, which was made available online in on October 8, 2025, and published in Volume 53, Issue 18 of the journal Nucleic Acids Research on October 14, 2025, details how they successfully recreated the RAN translation process in a mutated C9orf72 gene.
“Though there are previous studies on the repeat sequences associated with the neurodegenerative diseases, the role of non-canonical translation like RAN translation remains unexplored. Hence, we planned to understand the potential regulators of RAN using a bottom-up screening approach,” says Taguchi, as the motivation behind the study.
The researchers used the revolutionary human factor-based reconstituted translation system (human PURE), established by scientists from the University of Hyogo, Japan. The human PURE system can faithfully reproduce the C9orf72-mediated RAN(C9-RAN) translation process outside a cell by using highly purified human translation components. This enabled the team to focus only on the non-AUG initiation mechanisms without any interfering cellular side effects.
Through systematic screening, the researchers discovered that two canonical translation initiation factors, eIF1A and eIF5B, act as repressors of RAN translation. They confirmed that these factors enforce stricter control over non-canonical translation initiation at two distinct checkpoints. Specifically, eIF1A suppresses C9-RAN translation during the scanning phase by promoting the correct selection of the start codon, whereas eIF5B enforces translational control in the post-scanning phase, during the assembly of the large and small subunits of the ribosome. “The successful in vitro reconstitution of abnormal protein synthesis enabled the detailed elucidation of the mechanisms of action of eIF1A and eIF5B,” remarks Taguchi.
The researchers validated their findings in human cells, demonstrating that eIF1A and eIF5B repress RAN translation through independent and additive mechanisms. Notably, their experiments also shed light on the disease’s interaction with cellular stress. When cells are under stress, C9-RAN translation typically increases. However, they found that this increase was completely abolished when eIF1A was absent, highlighting its critical role in the so-called ‘integrated stress response-mediated enhancement’ of C9-RAN.
By pinpointing eIF1A and eIF5B as key regulators, this study sheds light on the intricate mechanisms behind the origin of ALS and FTD. “These newfound insights provide a foundation for developing technologies to control RAN translation, paving the way for novel therapeutic strategies against neurodegenerative diseases,” concludes Taguchi, hopeful about future research efforts.
Reference
- Authors:
- Hayato Ito1, Kodai Machida2, Yuzo Fujino3, Mayuka Hasumi1, Soyoka Sakamoto1, Yoshitaka Nagai3,4, Hiroaki Imataka2, and Hideki Taguchi1,5,*
*Corresponding author - Title:
- Canonical translation factors eIF1A and eIF5B modulate the initiation step of repeat-associated non-AUG translation
- Journal:
- Nucleic Acids Research
- DOI:
- 10.1093/nar/gkaf994
- Affiliations:
- 1School of Life Science and Technology, Institute of Science Tokyo, Japan
2Graduate School of Engineering, University of Hyogo, Japan
3Department of Neurology, Kindai University Faculty of Medicine, Japan
4Life Science Research Institute, Kindai University, Japan
5Cell Biology Center, Institute of Integrated Research, Institute of Science Tokyo, Japan
Related articles
Further information
Professor Hideki Taguchi
Institute of Integrated Research, Institute of Science Tokyo
- taguchi@bio.titech.ac.jp
Contact
Public Relations Division, Institute of Science Tokyo
- media@adm.isct.ac.jp
- Tel
- +81-3-5734-2975