How proteins bind to RNA: the dual mechanism of zinc fingers and disordered regions
Using molecular simulations, researchers uncover how disordered regions enhance specific RNA interactions in FUS protein-RNA complexes
RNA-binding proteins use a dual binding mechanism involving zinc finger (ZnF) domains and intrinsically disordered regions (IDR), reports a new study from Institute of Science Tokyo, Japan. Using advanced molecular modeling, the study analyzes a “FUS protein-RNA” complex—revealing how the protein uses its ZnF domain for RNA sequences recognition and its flexible IDR domain for its non-specific interactions. This breakthrough strategy is likely common to nucleic acid binding, offering fresh insights into molecular science.
Unlocking RNA Binding: How FUS Protein Uses Flexibility to Strengthen Precision
Kijima and Kitao (2025) | Journal of Chemical Information and Modeling | 10.1021/acs.jcim.5c01059
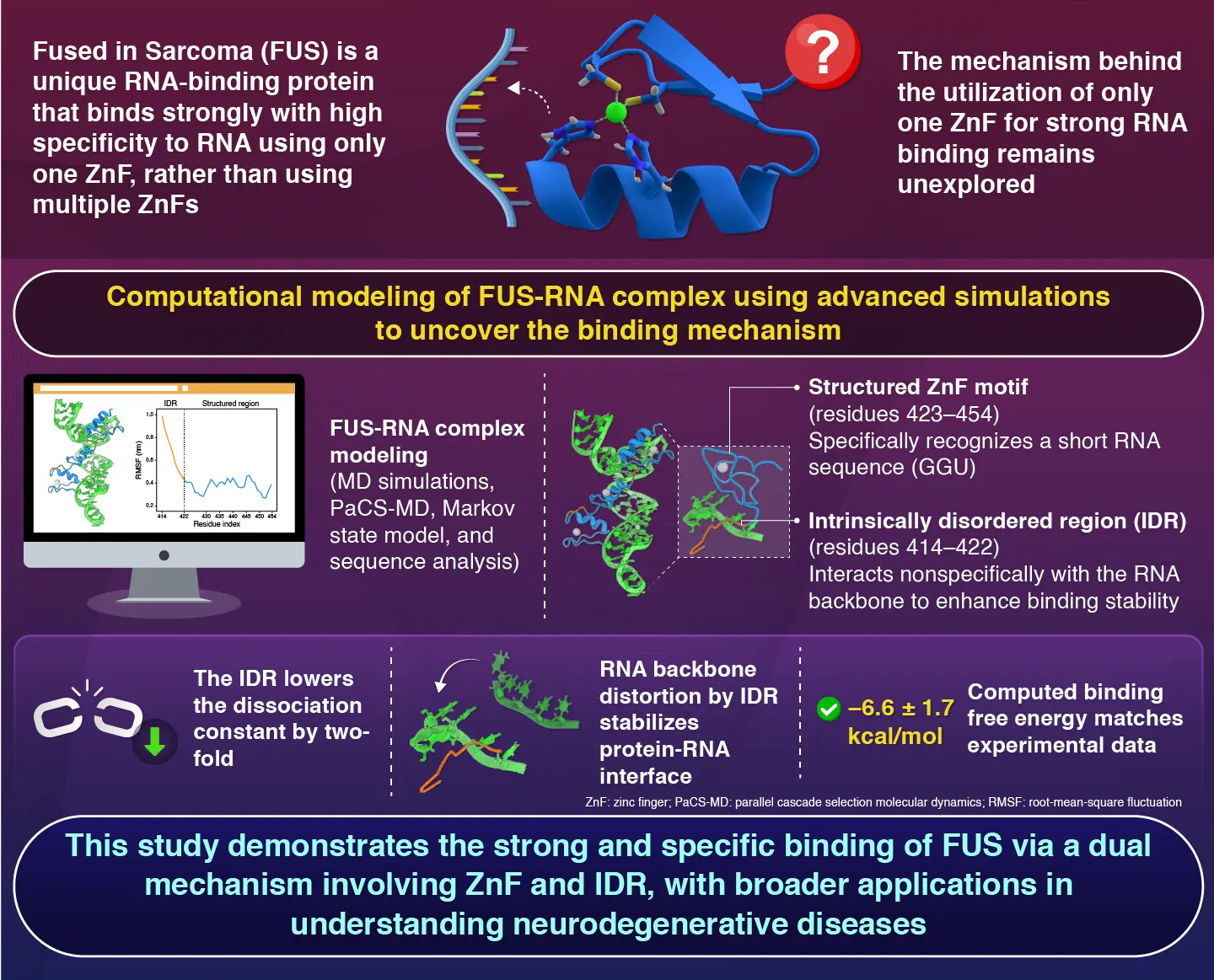
Proteins that can bind to nucleic acids play a critical role in various aspects of gene regulation, from DNA replication and repair to processing and RNA translation. Understanding how these proteins recognize and interact with the nucleic acids is the key to deciphering the underlying mechanism of how cells regulate gene expression and adapt to changing environments. While most of these proteins involve both well-structured domains and flexible, intrinsically disordered regions (IDRs), how these domains work together in protein-RNA binding remains unclear.
One notable example is the “Fused in Sarcoma” (FUS) protein, a well-known RNA-binding protein involved in gene regulation and neurodegenerative diseases. Although widely studied, its exact binding mechanism remains poorly understood. To overcome this gap, Professor Akio Kitao and PhD student Soichiro Kijima from the School of Life Science and Technology at Institute of Science Tokyo (Science Tokyo), Japan, employed molecular simulations to analyze the interactions between FUS protein and RNA sequences. The findings were made available online in the Journal of Chemical Information and Modeling on July 22, 2025, and published in Volume 65, Issue 15 on August 11, 2025.
The FUS protein has a distinct structure with a well-structured single-stranded RNA-binding RanBP2-type zinc finger (ZnF) domain followed by long disordered protein regions known as IDRs. ZnFs are small protein domains containing zinc ions and are the most common nucleic acid binding domains in eukaryotes. Using a combination of molecular dynamics and enhanced sampling techniques, the team simulated how the FUS protein interacts with a short RNA strand containing a known target sequence (GGU).
Their results revealed two distinct binding modes: one involving only the structured ZnF region and a second, more stable mode where both the ZnF domain and the disordered region interact with the RNA strand.
“We observed that the structured ZnF domain recognizes specific RNA sequences, but on its own, it binds only weakly,” explains Kitao. “Surprisingly, it was the disordered IDR which boosts this interaction.”
According to the findings, the disordered IDR region attaches to RNA in a non-specific way through charge-based interactions with phosphate groups, lowering the dissociation constant by two-folds. These interactions are independent of the sequences and enhance the overall binding affinity of the protein. Additionally, it was noted that these flexible IDR regions cause distortion of the RNA backbone which further stabilizes the protein-RNA interface. The combined effect of ZnF and IDR results in a more tightly held structure which is nearly 10 times stronger than ZnF alone.
Building on this, the researchers also conducted additional sequence analysis of other proteins containing IDRs, which suggested that this cooperative binding mechanism may be more common than previously known.
“Our findings suggest that IDRs aren’t just passive linkers,” notes Kitao. “They actively contribute to the RNA-binding mechanism and may have broader implications in molecular science.”
Overall, the study provides a new framework for mapping how nucleic acid binding proteins achieve both specificity and flexibility, which is critical in gene regulation. It also opens exciting avenues for drug design with dual-mode recognition for enhanced biomolecular sensing and gene-targeting. Looking ahead, the researchers plan to explore if this mechanism operates in other proteins involved in RNA metabolism and how post-translational modifications of disordered regions may affect binding dynamics—thereby deepening our understanding of the molecular mechanisms.
Reference
- Authors:
- Soichiro Kijima1 and Akio Kitao1*
- Title:
- RNA Binding Mechanism of the FUS Zinc Finger in Concert with Its Flanking Intrinsically Disordered Region
- Journal:
- Journal of Chemical Information and Modeling
- Affiliations:
- 1 School of Life Science and Technology, Institute of Science Tokyo, Japan
Related articles
Further Information
Professor Akio Kitao
School of Life Science and Technology, Institute of Science Tokyo
- akitao@life.isct.ac.jp
Contact
Public Relations Division, Institute of Science Tokyo
- media@adm.isct.ac.jp
- Tel
- +81-3-5734-2975