Catalytic mechanism of cPGA hydrolysis by Parkinson’s disease-linked DJ-1
The molecular mechanism of how DJ-1 hydrolyzes a reactive glycolysis side product reveals its pathophysiological significance in Parkinson’s disease
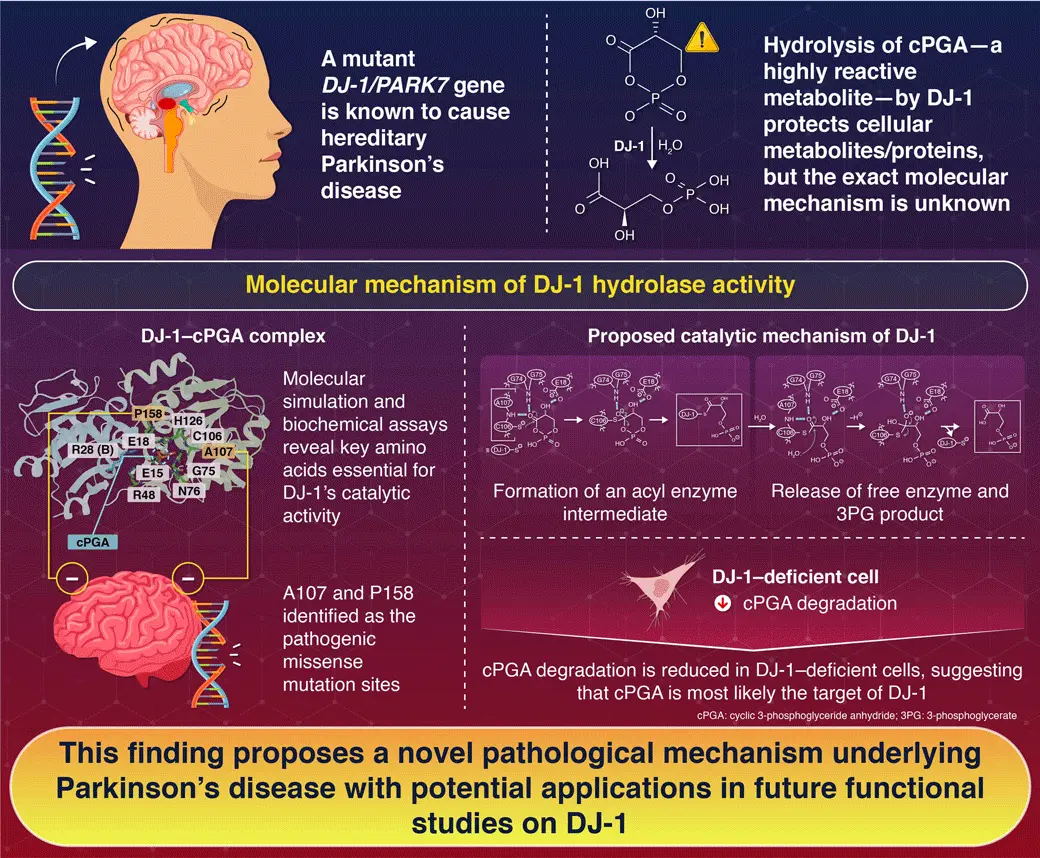
To understand how DJ-1 hydrolyzes cyclic 3-phosphoglyceric anhydride—a highly reactive, toxic cellular metabolite—researchers from Japan performed molecular simulations and biochemical assays, including mutational analyses, confirming DJ-1’s role in the pathogenesis of hereditary Parkinson’s disease. A mutant DJ-1 causes recessive Parkinson’s disease, but the molecular mechanism of this process has not been well studied. By revealing the amino acids involved in its catalytic activity, this work lays foundation for future functional studies on DJ-1.
Catalytic Mechanism Behind DJ-1 Hydrolysis of cPGA
Watanabe et al. (2025) | Journal of Cell Biology | 10.1083/jcb.202411078
DJ-1/PARK7, a gene linked to recessive familial Parkinson’s disease, encodes DJ-1, which has potential antioxidant activities, protecting cells from mitochondrial damage. With a range of biochemical roles attributed to it, including a redox-regulated chaperone, a transcriptional regulator, a glyoxalase, a cysteine protease, and a cyclic 3-phosphoglyceric anhydride (cPGA) hydrolase, the exact function of DJ-1 remains ambiguous. However, some facts about DJ-1 point towards the possibility of its primary role as a cPGA hydrolase. This enzymatic function fits aptly with DJ-1’s molecular structure, and DJ-1’s esterase activity reported earlier might reflect its role in cPGA hydrolysis. Owing to its instability, cPGA has been a difficult substrate to work with, limiting our understanding of DJ-1’s role in converting this highly reactive glycolysis side product to detoxified 3-phosphoglycerate (3PG).
To solve this puzzle, a collaborative team of researchers from Japan—under the leadership of Professor Noriyuki Matsuda and Associate Professor Yoshitaka Moriwaki from the Institute of Integrated Research, Institute of Science Tokyo (Science Tokyo), Japan—combined molecular simulations with biochemical assays and revealed the catalytic mechanism of cPGA hydrolysis by DJ-1. Their findings were published online in the Journal of Cell Biology on June 04, 2025. “Mutational analyses to identify the amino acid residues critical for cPGA hydrolase activity have been limited to C106, and no structural models of either the cPGA–DJ-1 complex or the hydrolytic mechanism have been proposed,” explains Matsuda about the motivation behind their study.
To demonstrate the molecular mechanisms behind cPGA hydrolase, the research team examined the structure of DJ-1 complexed with cPGA. The molecular dynamics simulations of this complex revealed the key amino acids involved in the binding pocket of DJ-1, critical for the identification and binding to cPGA. Next, they mutated these amino acid residues to elucidate the molecular mechanism of cPGA hydrolysis. These experiments revealed that E15 and E18 residues are important for the formation of the catalytic binding pocket and establishing hydrogen bonds with the cPGA molecule. G74, G75, and C106 residues participated in the stabilization and formation of a tetrahedral intermediate in the reaction pathway, whereas A107 and P158 residues determined the formation of hydrogen bonds with the molecular groups on cPGA and the formation of cPGA binding pockets, respectively.
Importantly, the researchers showed that the deletion of P158 and missense mutations in A107—also reported in hereditary Parkinson’s disease—completely revoked the cPGA hydrolysis activity of DJ-1 in vitro, confirming the pathophysiological implications of DJ-1. Based on these results, the research team proposed a new six-step molecular reaction model for DJ-1 hydrolase activity.
To examine the physiological relevance of DJ-1, the team compared cPGA hydrolysis in wild-type cells and DJ-1 knock-out cells. The cPGA hydrolase activity was significantly reduced in DJ-1-deficient cells, resulting in the accumulation of cPGA-modified metabolites in the cells. This suggests that among the reported substrates of DJ-1, cPGA is the primary physiological target and the mutations observed can result in the complete loss of cPGA-degrading function.
Summarizing the research team’s findings, Moriwaki and Matsuda conclude, “We believe that the molecular model presented in our research will provide solid insights for future functional studies of DJ-1 and enhance our understanding of the pathogenesis that leads to hereditary Parkinson’s disease.”
Reference
- Authors:
- Aiko Watanabe1, Shizuka Ogiwara1, Mirei Saito1, Masaki Mishima2, Masahiro Yamashina3, Ryuichiro Ishitani4, Yutaka Ito5, Keiji Tanaka6, Fumika Koyano1, Koji Yamano1,8, Hidetaka Kosako7, Yoshitaka Moriwaki4*, and Noriyuki Matsuda1*
- Title:
- The reaction mechanism for glycolysis side product degradation by Parkinson’s disease–linked DJ-1
- Journal:
- Journal of Cell Biology
- Affiliations:
- 1Department of Biomolecular Pathogenesis, Institute of Science Tokyo, Japan
2Department of Molecular Biophysics, Tokyo University of Pharmacy and Life Sciences, Japan
3Department of Chemistry, Institute of Science Tokyo, Japan
4Department of Computational Drug Discovery and Design, Institute of Science Tokyo, Japan
5Department of Chemistry, Tokyo Metropolitan University, Japan
6Laboratory of Protein Metabolism, Tokyo Metropolitan Institute of Medical Science, Japan
7Division of Cell Signaling, Tokushima University, Japan
8Intracellular Quality Control Project, Tokyo Metropolitan Institute of Medical Science, Japan
*Corresponding authors
Related articles
Further information
Professor Noriyuki Matsuda
Department of Biomolecular Pathogenesis, Institute of Science Tokyo
Associate Professor Yoshitaka Moriwaki
Department of Computational Drug Discovery and Design, Institute of Science Tokyo
Contact
Public Relations Division, Institute of Science Tokyo
- media@adm.isct.ac.jp
- Tel
- +81-3-5734-2975