How cells stay healthy: new insights into a selective protein cleanup system
Research reveals how GOMED identifies and removes unwanted proteins, highlighting therapeutic potential
A selective protein degradation system known as Golgi membrane-associated degradation (GOMED), which identifies and removes unwanted proteins, has been delineated by researchers at Science Tokyo. This system works by tagging problematic proteins with a “molecular label” called K33-linked ubiquitin and using an adaptor protein, optineurin (OPTN), to guide them to GOMED structures for breakdown. These findings improve our understanding of cellular self-cleanup processes and may help in developing new treatments for neurodegenerative diseases.
How Cells Use GOMED to Choose and Destroy Unwanted Proteins
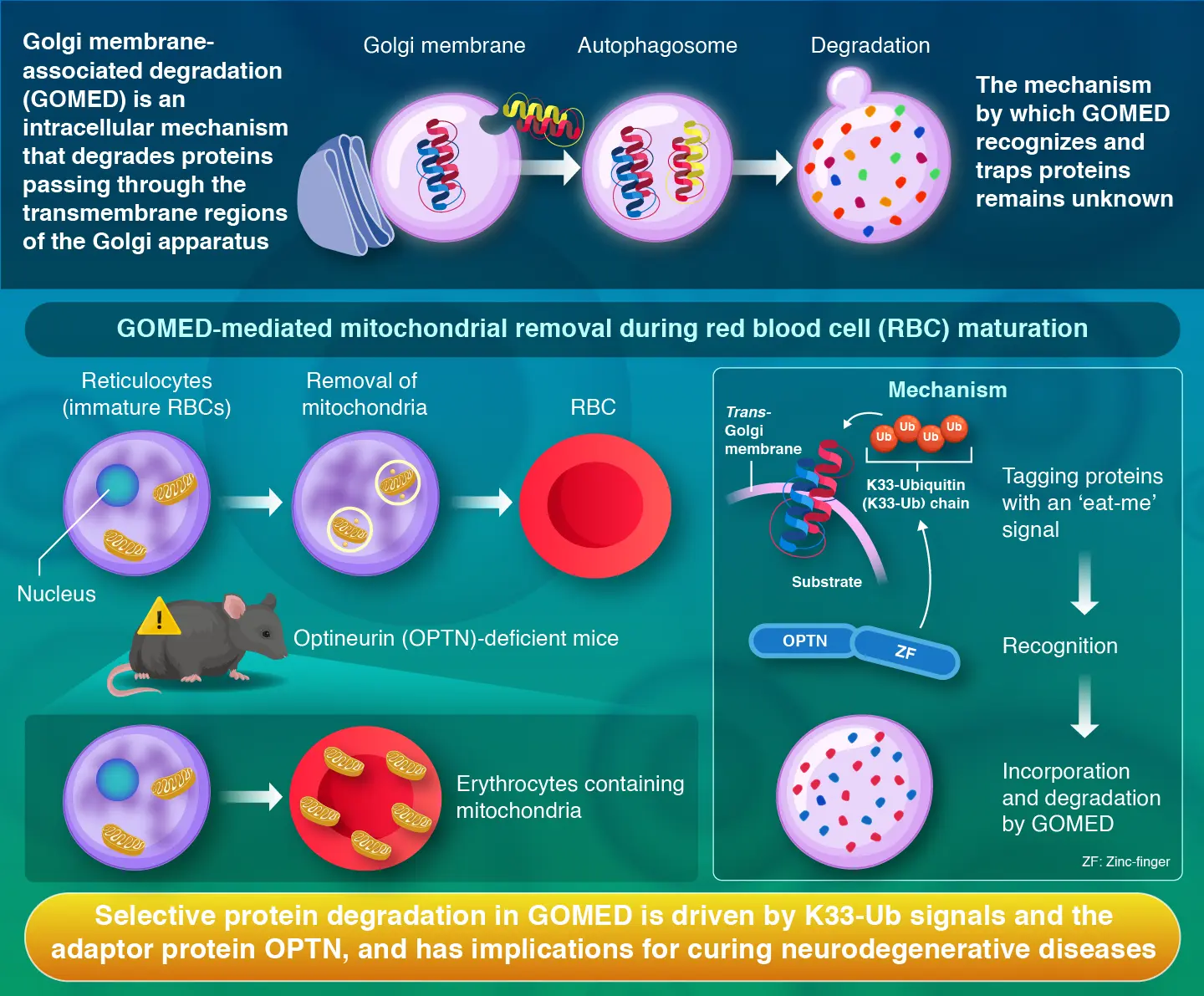
To stay healthy, our cells rely on a self-cleaning mechanism that removes defective or unnecessary components. This process, known as autophagy, has been linked not only to cellular maintenance but also to various diseases and aging. Recently, a related process called Golgi membrane-associated degradation (GOMED) was discovered, which degrades proteins that pass through the transmembrane region of the Golgi apparatus. GOMED closely resembles autophagy, in which targets are engulfed inside a double membrane and then broken down by lysosomes. While the existence and general functions of GOMED are known, the way it recognizes which proteins to target has remained unclear.
Now, new findings by researchers at Institute of Science Tokyo (Science Tokyo), Japan, reveal how proteins are marked for GOMED, providing fresh insights into this long-standing question. The study was led by Project Assistant Professor Yoichi Nibe-Shirakihara and Distinguished Professor Shigeomi Shimizu from Science Tokyo, in collaboration with Hiroshima University, University of Melbourne, and Monash University. Their study was published online in Volume 16 of the journal Nature Communications on October 20, 2025.
“The molecular dynamics and functions of GOMED are being clarified, but how GOMED recognizes and degrades its substrates has not been elucidated to date. Therefore, we investigated whether GOMED degrades substrates selectively and explored its molecular mechanism,” explains Nibe-Shirakihara.
To begin their search, the scientists looked for adaptor proteins that might guide GOMED toward its targets. They focused on several adaptors already known from autophagy research and studied them in genetically modified cells that lacked normal autophagy, ensuring that any degradation they observed was due specifically to GOMED.
When the cells were exposed to a GOMED stress-inducing drug, only one adaptor protein, optineurin (OPTN), showed clear changes. Its levels first increased and then decreased when lysosomal degradation was blocked, suggesting that OPTN plays a direct role in the GOMED pathway.
In autophagy, OPTN is known to interact with ubiquitin, a small molecular tag that marks proteins for destruction, and deliver these tagged proteins to autophagosomes, which are double-membrane structures, where degradation occurs. Based on this knowledge, the researchers hypothesized that OPTN might play a similar role in GOMED.
They tested this idea using a model protein called vesicular stomatitis virus G protein VSV-Gt045 (VSVG)-GFP, which is commonly used to study how proteins move through the Golgi apparatus. Under conditions of Golgi stress, this protein was tagged with a specific form of ubiquitin known as K33-linked polyubiquitin. The team discovered that this K33-linked polyubiquitin tag acts as a unique ‘eat-me’ signal for GOMED. OPTN recognizes this signal through its zinc-finger domain and helps deliver the tagged proteins to GOMED-related structures for degradation.
These findings were confirmed in animal experiments. In mice lacking OPTN, mitochondria were not properly removed from developing red blood cells, demonstrating that this mechanism is essential to living organisms. “We found that selective protein degradation occurs in GOMED through the recognition of substrates using K33-linked ubiquitin chains as an ‘eat-me’ signal, with OPTN acting as an adaptor molecule,” says Nibe-Shirakihara.
By uncovering how GOMED identifies and destroys its targets, this study fills a major gap in our understanding of cellular quality control processes. Because GOMED is implicated in neurological disorders and other diseases, this knowledge could eventually lead to new therapeutic strategies to restore proper cellular cleanup, slow down disease progression, or prevent the toxic buildup of damaged proteins, offering hope for treating a range of currently incurable disorders.
Reference
- Authors:
- Yoichi Nibe-Shirakihara1*, Shinya Honda1, SatokoArakawa1, SatoruTorii1, Hajime Tajima Sakurai1, Hirofumi Yamaguchi1, Shigeru Oshima2, Ryuichi Okamoto2, Michael Lazarou3,4,5, Hideshi Kawakami6, and Shigeomi Shimizu1*
*Corresponding authors - Title:
- Optineurin is an adaptor protein for ubiquitinated substrates in Golgi membrane-associated degradation
- Journal:
- Nature Communications
- Affiliations:
- 1Department of Pathological Cell Biology, Advanced Research Initiative, Institute of Integrated Research, Institute of Science Tokyo, Japan
2Department of Gastroenterology and Hepatology, Graduate School,Institute of Science Tokyo, Japan
3Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Australia
4Walter and Eliza Hall Institute of Medical Research, Australia
5Department of Medical Biology, University of Melbourne, Australia
6Department of Epidemiology, Research Institute for Radiation Biology and Medicine, Hiroshima University, Japan
Related articles
Further information
Project Assistant Professor Yoichi Nibe
Department of Pathological Cell Biology, Advanced Research Initiative, Institute of Integrated Research, Institute of Science Tokyo
- ynibe.pcb@tmd.ac.jp
Distinguished Professor Shigeomi Shimizu
Department of Pathological Cell Biology, Advanced Research Initiative, Institute of Integrated Research, Institute of Science Tokyo
Contact
Public Relations Division, Institute of Science Tokyo
- media@adm.isct.ac.jp
- Tel
- +81-3-5734-2975